Introduction
Data science applications (such as the ones built with Shiny, Dash, Streamlit, Gradio, etc.) produced by researchers are considered to be research output just like more traditional research outputs such as journal articles, conference papers, book chapters, etc. This means that when creating and publishing such applications researchers need to fulfil funder and institutional requirements.
For example, researchers need to ensure that their applications fall within the existing ethical approval in projects where such approval is applicable (see guidelines from e.g., Swedish Research Council, ERC). In addition, they need to meet open access publication requirements (e.g., Swedish Research Council, ERC), open data requirements (e.g., Swedish Research Council, ERC), open analysis workflows and code requirements (e.g., National guidelines for promoting open science in Sweden).
To meet these requirements, European and Swedish funders and universities currently recommend adhering to FAIR principles (e.g., Swedish Research Council: Making research data accessible and FAIR criteria).
What is FAIR
FAIR (Findable, Accessible, Interoperable, Reusable) is a set of principles originally written for research data (see Wilkinson et al 2016) but since expanded to other research output (see Baker et al 2022, Patel et al 2023). There is no one specific way to 'make something FAIR'; instead, research output can adhere to FAIR principles to different extent and in different ways.
The long-term goal of SciLifeLab Serve is allow Swedish researchers to meet funder requirements in terms of FAIR principles when publishing applications without any extra work; in other words, everything should be done for you automatically when you publish your app on SciLifeLab Serve. In the meantime, there are some things that researchers can do themselves. On this page, we give recommendations on basic steps researchers can take to adhere to FAIR principles to a reasonable extent when sharing data science applications.
Meeting FAIR requirements
A data science application consists of research data and code that is used to build the application. Therefore, you need to think about FAIR sharing of both data and code.
Deposit your data and code in a repository
The easiest way to adhere to most of the FAIR requirements is to deposit data and code to a repository that complies with FAIR requirements. This means that the repository will, for example, issue a persistent identifier for your data and code (e.g., a Digital Object Identifier, DOI), keep track of different versions, commit to long-term availability and archiving, make the information about your data and code available in an interoperable way (e.g., through an API interface in JSON format), etc.
One example of such a repository is Zenodo. Zenodo is a general purpose repository run by CERN with funding from the EU funding programmes. Zenodo has a page where they describe exactly how they comply with FAIR requirements. There are many other repositories fulfilling FAIR requirements that are, for example, aimed at a specific data type, at a specific scientific discipline, or at researchers from specific institutions. For example, in Sweden the following universities have own repositories: Karolinska Institutet, KTH Royal Institute of Technology, Stockholm University.
If you are unsure which repository to use, you can try searching in the Registry of Research Data Repositories and asking the data management support in your own university or the national helpdesk organised by SciLifeLab (data-management@scilifelab.se).
When you identify the most suitable repository for your case, upload your code and data to that repository and make it available. There is however a way to automate it (see the next section).
In practice: GitHub
When you share a data science application, in practice the easiest and most pragmatic option will be to deposit your code and data to GitHub. GitHub is easy because most researchers either already know how to use it or plan to learn how to use it. We encourage you to share your code and data this way but unfortunately that is not yet sufficient to meet FAIR requirements. This is because GitHub does not guarantee availability and does not issue persistent identifiers (e.g., a GitHub release is easy to overwrite).
Once your code and data are on GitHub there is however a relatively easy way forward. You can, for example, use the Zenodo integration with GitHub - see the documentation about Zenodo here. First, you will need to connect your Zenodo account with your GitHub account. Subsequently, you will see a list of your GitHub repos through the Zenodo interface, and you can activate automatic archival for the repo that you are interested in. From then on, every time you do a GitHub release the content of your GitHub repo will automatically be deposited on Zenodo.
Once your code and data is on Zenodo through the GitHub integration you will automatically meet all the minimaal FAIR requirements. For example, you will have a DOI for your code and data, there will be specific versions that cannot be overwritten, etc. You can see an example of one of our own apps here: GitHub repo, Zenodo entry.
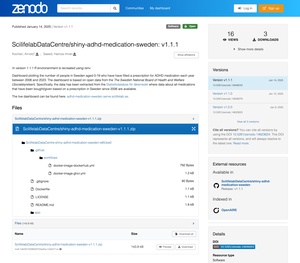
Depositing research data separately
While it is good to make data available together with your application code, often there is a data repository where the researcher community will expect to find your data. In these cases we recommend depositing your data into that repository as well. This way, even if someone does not know about your application they can still find your dataset when they are searching in appropriate data catalogues.
Another case where you would need to deposit your data separately is if your data files are too big for GitHub (at the moment GitHub does not allow files larger than 100 MiB). In these cases we recommend depositing the data in a dedicated or general data repository and fetching it in your application using a persistent URL from that repository. For example, you can deposit the data as its own record on Zenodo and download these data when you launch your app. If you need help with this do not hesitate to get in touch with us for assistance - serve@scilifelab.se.
Going the extra mile: FAIR sharing of Docker images
Sharing your code and data through GitHub and Zenodo or similar as described above is in priciple sufficient to say that you meet the FAIR requirements. However, those who wish can go the extra mile and improve on that - deposit the Docker image with the application as well.
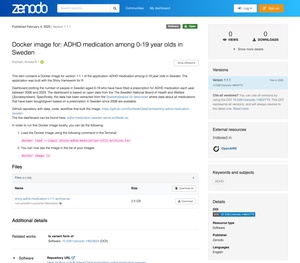
In order to host your application on SciLifeLab Serve, you will have created a Docker Image and published it in a public image registry, e.g., DockerHub or Github Container Repository. Just like in case of code and data on GitHub, these public image registries are not yet sufficient to meet FAIR requirements. They do not guarantee availability, can be overwritten, etc. In this case too, the recommendation is to deposit your Docker image to a repository that facilates meeting FAIR requirements. This can also be Zenodo or a similar repository.
Docker images are archives that are not visible as files on your computer by default but can be saved as files if needed. To do that, run the command docker save image/name:tag -o image-name.tar in the folder where you want to save it. The resulting .tar file can be uploaded to Zenodo or any other repository. See an example of a Zenodo entry with a Docker image here.
Structure and describe your data and code
One of the important pillars of FAIR is presence of good metadata - good descriptions of your data and code that will make it easier to understand what you did, why, and how it can be reused. Structure of code or data is also important to make sure other members of the community can quickly and easily navigate what you shared. Therefore, where possible, community standards for structure and descriptions should be used.
Various data types will have different community standards for formats, data structure, and expected descriptions that you can adhere to. The easiest way to find out what you should adhere to is imagine what you yourself as a researcher in that field would expect. You can also look online for a standard in your particular scientific field or for your particular data type. You can also ask the data management support at your university or the national helpdesk organised by SciLifeLab (data-management@scilifelab.se) for tips.
When it comes to describing your code, there is no single community standard but you should make sure to adhere to standard practices (e.g., install your Python packages using a requirements.txt file or similar rather than in the scripts). Adhere to the conventions for the specific frameworks that you use (e.g., naming and structuring of scripts suggested in the official documentation). It is also a good idea to adhere to good coding practices and leave useful comments in the code. Importantly, add a README file (instructions from GitHub) file describing the structure of your project/code and provide a set of example commands on how one can launch the application.
Specify a license
The license specifies the terms and conditions under which your app data or code can be used and distributed. When there is no specific license attached to the data and code of your application, the default assumption will be that they should not be distributed or reused at all to avoid any conflicts.
There are different liceses for code and research data, and you should make sure to use appropriate licenses (e.g. license for software should not be applied to data). For code, we recommend specifying one of the software licenses approved by the Open Source Initiative - see the list of licenses here; the MIT license is a popular choice that is a great default. For research data you can use one of the Creative Commons licenses; here CC BY 4 is a popular choice that is a great default.
The easiest way to attach a license is to create a LICENSE file (see instructions on GitHub) with your code and a license page in your application (see an example of such a page here).
Lock the versions
Another important aspect of meeting FAIR requirements is ensuring reproducibility - someone who launches your application years later should still get the same result. One easy practice is to always use specific versions of languages and packages. For example, specify which specific version of Python you use and specify which version of packages should be installed in your code rather than just installing the latest. If you are creating a Shiny app with R we recommend using renv to save and recreate the specific environment (this is described in our user guide for Shiny applications).
Needless to say, make sure to always specify which version of the data each application version relies on.
Let others know how to cite your code
If others are going to reuse your application they should also cite it in their publications so that you get some credit for it. To let others know how to correctly cite your application you can include a CITATION.cff file. This file format is a convention integrated by both GitHub and Zenodo. Read more about CITATION.cff here.
Other sources of information
Here we wrote guidelines from the perspective of typical use cases of SciLifeLab Serve but there are many other good sources of information about FAIR that you can use if you are interested to dive into this. Here are some recommendations:
FAIR Biomedical Research Software (FAIR-BioRS) Guidelines
University of Groningen: FAIR research software
EVERSE: Tools and practices for FAIR research software development
The SciLifeLab Serve user guide is powered by django-wiki, an open source application under the GPLv3 license. Let knowledge be the cure.


